Members
・応募書類の受付をした応募者にメールで通知いたしました(通知方法を変更したため、マイページではご確認いただけません)。メールが届いていない場合は recruit@mail2.adm.kyoto-u.ac.jp までご連絡下さい。
・書類選考の結果については9月下旬頃にメールにてご連絡をさせていただく予定です。
・選考スケジュールの変更がありましたら、ホームページ上でご連絡いたします。
-We sent an email to the applicants whose proposals had been received. (Notification method has been changed. We will not notify you on your applicant’s personal page.) If you have not received the email, please contact to recruit@mail2.adm.kyoto-u.ac.jp.
-We will inform you of the results of the document screening by e-mail around late-September.
-If we change any screening schedule, we will inform you on our website.
14th
Term: Apr. 2024 ~
 :Global Type
:Global Type
 :Tenure-track Type
:Tenure-track Type
● YAMADA Shintaro Assistant Professor
Research Interests: DNA recombination and repair
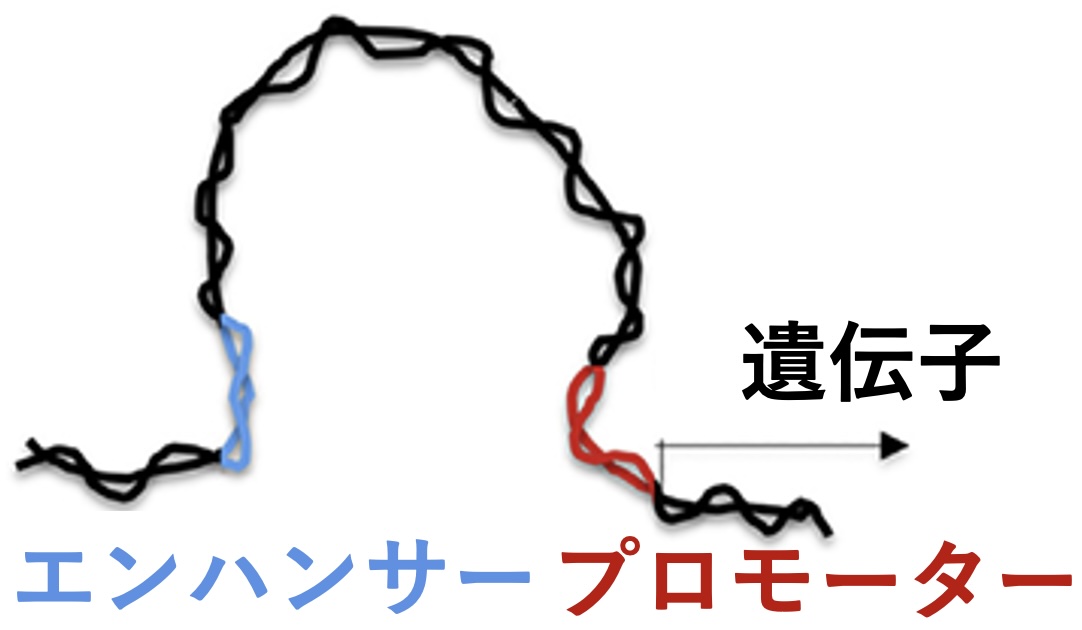
Research Topic: Identifying carcinogenesis-related transcriptional regulatory elements (enhancers) comprehensively and elucidating the mechanism in hormone responsive oncogene overexpression and tumor formation in DNA repair deficient cancer models for understanding cancer development fundamentally
Host Department: Graduate School of Medicine
Previous Affiliation: Graduate School of Biostudies, Kyoto University
Short Introduction
Recent advances in next-generation sequencing technology have expanded targets of mutation analysis from individual genes to the whole genome. The whole genome mutation analysis is expected to help uncover new mechanisms in cancer development, which may lead to development of new cancer therapies. Nevertheless, it is not easy to identify cancer-causing mutations. This is because those mutations are often detected together with a large number of mutations in junk sequences, which do not directly drive cancer development. This study investigates enhancers, i.e., genomic regions important for transcriptional regulation of genes. It has been suggested that there are many unknown enhancers in the human genome. Colleagues and I have been working on the identification of novel enhancers, with which I will focus on enhancer mutations in the whole genome mutation analysis. The updated enhancer map serves as a basis for development of a new method to eliminate junk regions from analysis. Outcomes of this study will contribute to precision, or personalized, medicine for cancer based on individual genetic information. Because enhancers are involved in cell-type specific transcriptional controls of genes, the findings will also help elucidate the organ specificity of cancer development.








